Sample Filters
Vizome allows you to work with the entire set of samples in our BeatAML dataset by default.

But you may want to apply a filter so that you're working with only a subset of the samples.
Vizome offers four different ways to apply sample-based filters. They have the following labels: Bar charts, Serial, List, and Genetics.
Click on a small bar graph to open a larger version of it. In the opened chart, click individual bars to select/de-select samples.
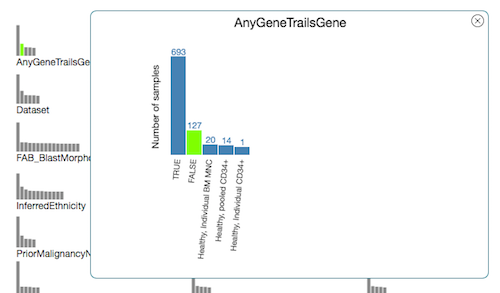
If you want to add another category, you may decide whether to combine selections across categories with "or" or "and." Note that within the same category, selections will always be combined with "or." It is only across categories that the combination method can change.
When you have chosen your desired subset of samples, give it a label, if desired, and click  and the subset will be the only samples that appear throughout Vizome for the current browsing session.
and the subset will be the only samples that appear throughout Vizome for the current browsing session.
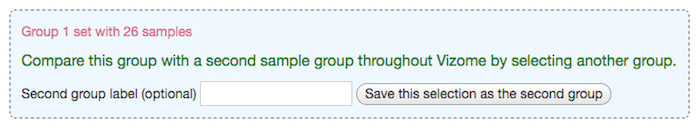
After saving one group, you will have the option to repeat the steps to save another group, in order to compare two groups of samples across Vizome. Note that the two groups must be composed of different samples. If any samples in the second group are already included in the first group, the second group cannot be saved.
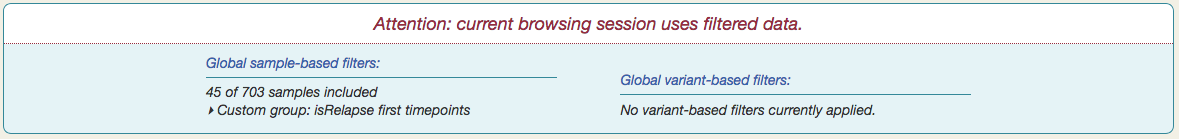
You will see a notification appear at the top of the page, summarizing the current set of global filters.
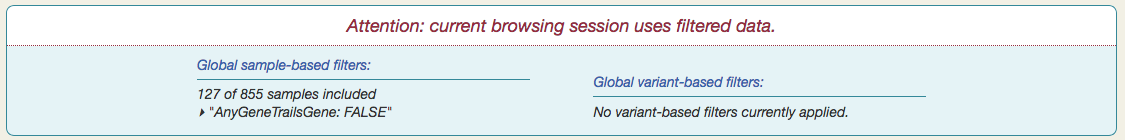
The Serial filtering option gives you fine-grained control over which samples make up your subset. When you select a category — isRelapse, for example — sample data appears in tabular form, with "Timepoints" (one per sample) as columns and patients as rows.
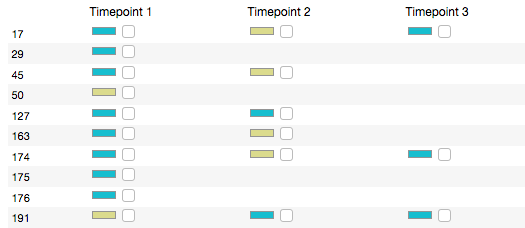
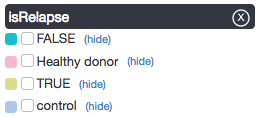
Values for each sample appear as color-coded rectangles with a checkbox. You can select or unselect all samples with a certain value — TRUE, for example — via the checkboxes in the legend at right.
If you want to add another category, you may decide whether to combine selections across categories with "or" or "and." Note that within the same category, selections will always be combined with "or." It is only across categories that the combination method can change. You may select/unselect individual samples at any time by checking or unchecking the box in the sample's table cell.
Below the table, you will see a count of the total number of samples selected. When you have chosen your desired subset of samples, give it a label, if desired, and click  . The subset will then be the only samples that appear throughout Vizome for the current browsing session.
. The subset will then be the only samples that appear throughout Vizome for the current browsing session.
You will see a notification appear at the top of the page, summarizing the current set of global filters.
If you already know the IDs (e.g., "17-00123") of the samples you would like to use as a subset, the List filtering option gives you an easy way to set that up.
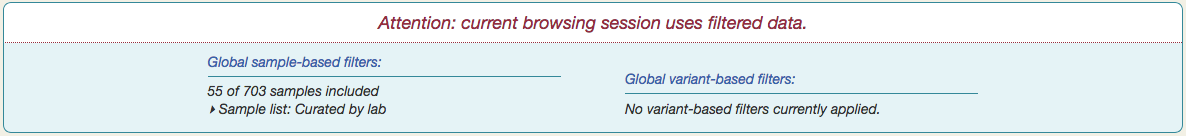
Enter a list of sample IDs separated either by commas or line breaks. Give the list a label, if desired, and click  . The subset will then be the only samples that appear throughout Vizome for the current browsing session.
. The subset will then be the only samples that appear throughout Vizome for the current browsing session.
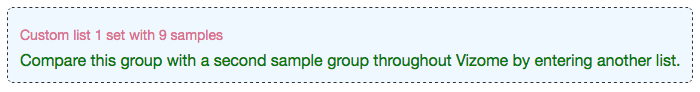
After saving one group, you will have the option to enter a new list of sample IDs to save another group, in order to compare two groups of samples across Vizome. Note that the two groups must be composed of different samples. If any of the samples in your second group are already included in your first group, the second group cannot be saved.
You will see a notification appear at the top of the page, summarizing the current set of global filters.
The Genetics filtering option allows you to define a subset of samples based on their shared genetic traits. For example, you may want to work with all samples that have both a variant in NPM1 and the internal tandem duplication in FLT3 (which appears in Vizome as FLT3_ITD). To begin, you would select NPM1 from the Gene drop-down menu. This will populate a table with all the unique variants in NPM1.
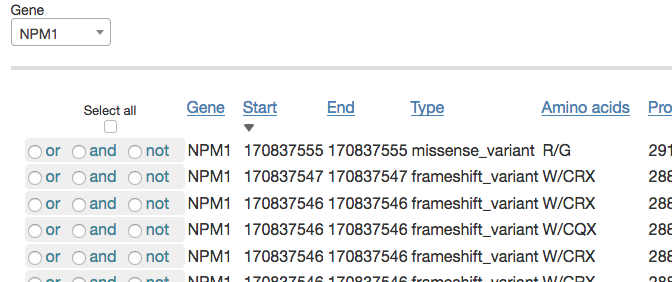
You would then repeat the gene search step to add FLT3_ITD, and click the box under "Select all." All samples with any variants in that table will be selected. If you then change the radio button for FLT3_ITD to "and" instead of "or," you should see the total number of samples decrease from 212 to 58, as that changes the selection to require that samples have both the FLT3 internal tandem duplication and a variant in NPM1.
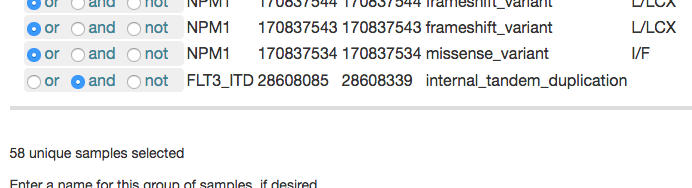
You could then give the subset a label, if desired, and click  . The subset will then be the only samples that appear throughout Vizome for the current browsing session.
. The subset will then be the only samples that appear throughout Vizome for the current browsing session.
After saving the subset, a notification appears at the top of the page, summarizing the current set of global filters.
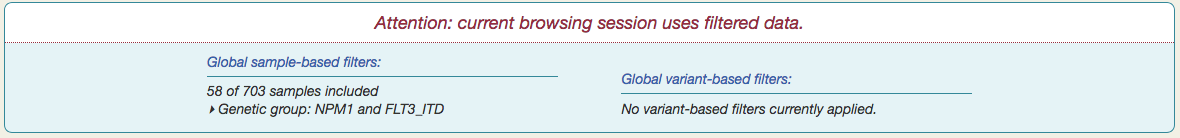

 and the subset will be the only samples that appear throughout Vizome for the current browsing session.
and the subset will be the only samples that appear throughout Vizome for the current browsing session.













 . The subset will then be the only samples that appear throughout Vizome for the current browsing session.
. The subset will then be the only samples that appear throughout Vizome for the current browsing session.






